In general, thermal shift assays can be applied to understand the thermal stabilization that proteins undergo when binding to a ligand. Thermal shift assays have been used in the past to understand proteins and their interactions, often with a focus on drug development. The cellular thermal shift assay (CETSA) takes this a step further by focusing on specific interactions of drug candidates within the cell.
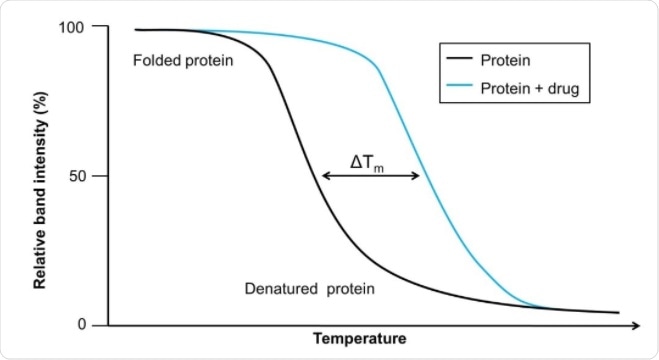
Image Credit: https://www.sygnaturediscovery.com/drug-discovery/bioscience/biophysical-assays/cellular-thermal-shift-assay-cetsa/
Why CETSA can be needed
The efficacy of a drug depends on how well they bind to their target, which is often a protein. If the drug binds to the target excessively or binds to other molecules, there can be significant adverse effects. These effects, caused by excessive or non-specific binding, can be difficult to predict in vitro because efficacy depends on the effective drug concentration and by factors that influence the conformation of proteins.
The issue in studying these possible drug targets was that the direct binding could not be monitored. Rather, cellular processes occurring downstream of binding were measured. In many cases, this meant that drugs failed in clinical trials because they did not bind the predicted cellular target. CETSA bypasses this by making use of shifts in thermal stability that occur when drugs bind to proteins.
The CETSA was developed in 2013 but has been widely used since then. Unlike its predecessors, CETSA can be used to confirm the link between an observed phenotypic response to a drug with a specific molecular interaction.
How does CETSA work?
Thermal shifts indicate the extent of unfolding in proteins. Existing thermal shift assays have been used to show that thermal shifts correlate with median inhibitory concentration values. However, these are only used on purified proteins and therefore miss out on the cellular context.
CETSA uses cellular contents rather than only purified proteins. The contents of the cell are heated to different temperatures, which leads to the denaturation and precipitation of proteins that are not bound to a ligand. The cell is then lysed so that the cell contents can be analyzed. Adding drugs to these cellular mixtures alters the thermal melting curve of the proteins as more become bound to the drug.
This is followed by the separation of the soluble fraction from the precipitated proteins. The soluble fraction contains the ligand-bound proteins, thus allowing for the quantification of the target protein remaining in the soluble fraction.
Detection of the stable protein can be done using quantitative western blotting or using antibodies. For western blots, the cell debris is separated from the precipitate. For the antibody approach, native proteins are identified using antibody pairs. Recent developments also allow for CETSA to be carried out on intact cells and tissues.
Rather than changing the temperature, CETSA can also be used to investigate the effects of different drug concentrations by keeping the temperature constant and altering the drug doses. This is called isothermal dose-response fingerprint (ITDRFCETSA). This results in a clear picture of the engagement of the drug to the target protein along the drug concentration axis.
There are also new developments to increase the applicability of CETSA. By combining CETSA with a split Nano Luciferase approach, the method can be used to engage targets for applications that are medium and high throughput. This improved methodology also allows for the screening of targets where phenotypic or other assays are not available, thereby expanding the range of CETSA.
Uses of CETSA
While relatively new, CETSA has been used on several targets already. The usable output from a CETSA experiment are melting curves for when the protein’s ligand is present or absent, which can then be compared across a panel of temperatures.
CETSA was initially validated using antifolate cancer drugs methotrexate and raltitrexed. These drugs target the proteins dihydrofolate reductase and thymidylate synthase, which showed big thermal shifts when tested with and without their drugs in CETSA.
Following its validation, CETSA was applied to test the specificity of a cancer drug in clinical development. The drug, called PD0332991, was believed to target cyclin-dependent kinases (CDKs), and the use of CETSA was able to show that the drug is selective for CDK4 and CDK6, and did not bind other CDKs such as CDK2 or CDK9.
Sources
- Molina, D., Jafari, R., Ignatushchenko, M., Seki, T., Larsson, E., Dan, C., Sreekumar, L., Cao, Y., and Nordlund, P., 2013. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science, 341(6141), pp. 84-87.
- Jafari, R., Almqvist, H., Axelsson, H., Ignatushchenko, M., Lundbäck, T., Nordlund, P., and Molina, D., 2014. The cellular thermal shift assay for evaluating drug target interactions in cells. Nature Protocols, 9(9), pp. 2100-2122.
- Martinez, N., Asawa, R., Cyr, M., Zakharov, A., Urban, D., Roth, J., Wallgren, E., Klumpp-Thomas, C., Coussens, N., Rai, G., Yang, S., Hall, M., Marugan, J., Simeonov, A. and Henderson, M., 2018. A widely-applicable high-throughput cellular thermal shift assay (CETSA) using split Nano Luciferase. Scientific Reports, 8(1).
Last Updated: Dec 2, 2020

Written by
Sara Ryding
Sara is a passionate life sciences writer who specializes in zoology and ornithology. She is currently completing a Ph.D. at Deakin University in Australia which focuses on how the beaks of birds change with global warming.
Source: Read Full Article
