During the present coronavirus disease 2019 (COVID-19) pandemic, multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOC) have emerged, including the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) variants. All VOCs are associated with increased virulence, transmissibility, and/or resistance to neutralizing antibodies in vaccinated or previously infected individuals.
 Study: SARS-CoV-2 Beta variant infection elicits potent linage-specific and cross-reactive antibodies. Image Credit: Juan Gaertner / Shutterstock.com
Study: SARS-CoV-2 Beta variant infection elicits potent linage-specific and cross-reactive antibodies. Image Credit: Juan Gaertner / Shutterstock.com
The VOCs have a variety of mutations within the SARS-CoV-2 spike (S) protein, several of which are present within the receptor-binding domain (RBD). Some mutations result in enhanced binding to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell. One example of this type of mutation is the mutation N501Y, which is linked to increased transmissibility of SARS-CoV-2. However, with immunity acquired through infection or via vaccination, mutations might arise more closely linked to antibody escape.
Several studies have examined RBD antibodies in COVID-19 patients before variants of SARS-CoV-2 were identified, which are termed non-VOC antibodies. Non-VOC RBD antibodies have been found to exhibit a select response for specific epitopes with enhanced recruitment of certain germline genes, with the most prominent being VH3-53 and closely related VH3-66, as well as VH1-2. One recent study assessed the serum of Beta variant infected patients to determine whether major non-VOC antibodies could neutralize this virus.
What is the difference in reactivity between RBD Beta and non-VOC RBD antibodies?
The authors of the current study, posted to the bioRxiv* preprint server, isolated CD19+CD27+ memory B-cells from 12 participants who had SARS-CoV-2 Beta variant infections to investigate the difference in reactivity between RBD Beta and non-VOC RBD on the level of monoclonal antibodies (mAbs). When compared to mAbs from non-infected individuals, sequence analysis from the participants revealed enrichment of certain VH genes, such as VH1-58, VH3-30, VH4-39, and VH3-53, which shows preferential recruitment of VH genes.
Within the mAbs taken from the cohort of this study, the number of somatic hypermutations was relatively low. This finding is consistent with previous reports on non-VOC SARS-CoV-2 infections. Collectively, the findings highlight the conservation of specific antibody sequence features between different antibody responses within individuals and between antibody responses that arise against the SARS-CoV-2 Beta variant and non-VOCs.
The authors compared antibody sequences following Beta infection to all non-VOC antibodies that had previously been published. To this end, the authors discovered several clonotypes, some of which were present in multiple participants of the study. These results show that an antibody subset to RBD Beta and non-VOC RBD converge on the recruitment of specific germline genes.
What are the binding properties of antibodies elicited by the SARS-CoV-2 Beta variant?
To establish the binding properties of antibodies elicited by the SARS-CoV-2 Beta variant, the authors selected mAbs for expression and further characterization based on specific criteria. These criteria included mAbs that were clonally expanded in one patient, mAbs of clonotypes present in several participants to interpret shared antibody response to RBD Beta, mAbs of clonotypes to identify cross-reactive mAbs, VH3-53/VH3-66 mAbs to interpret the unexpected recurrent shared antibody response against RBD Beta, and mAbs of VH genes with the strongest enrichment.
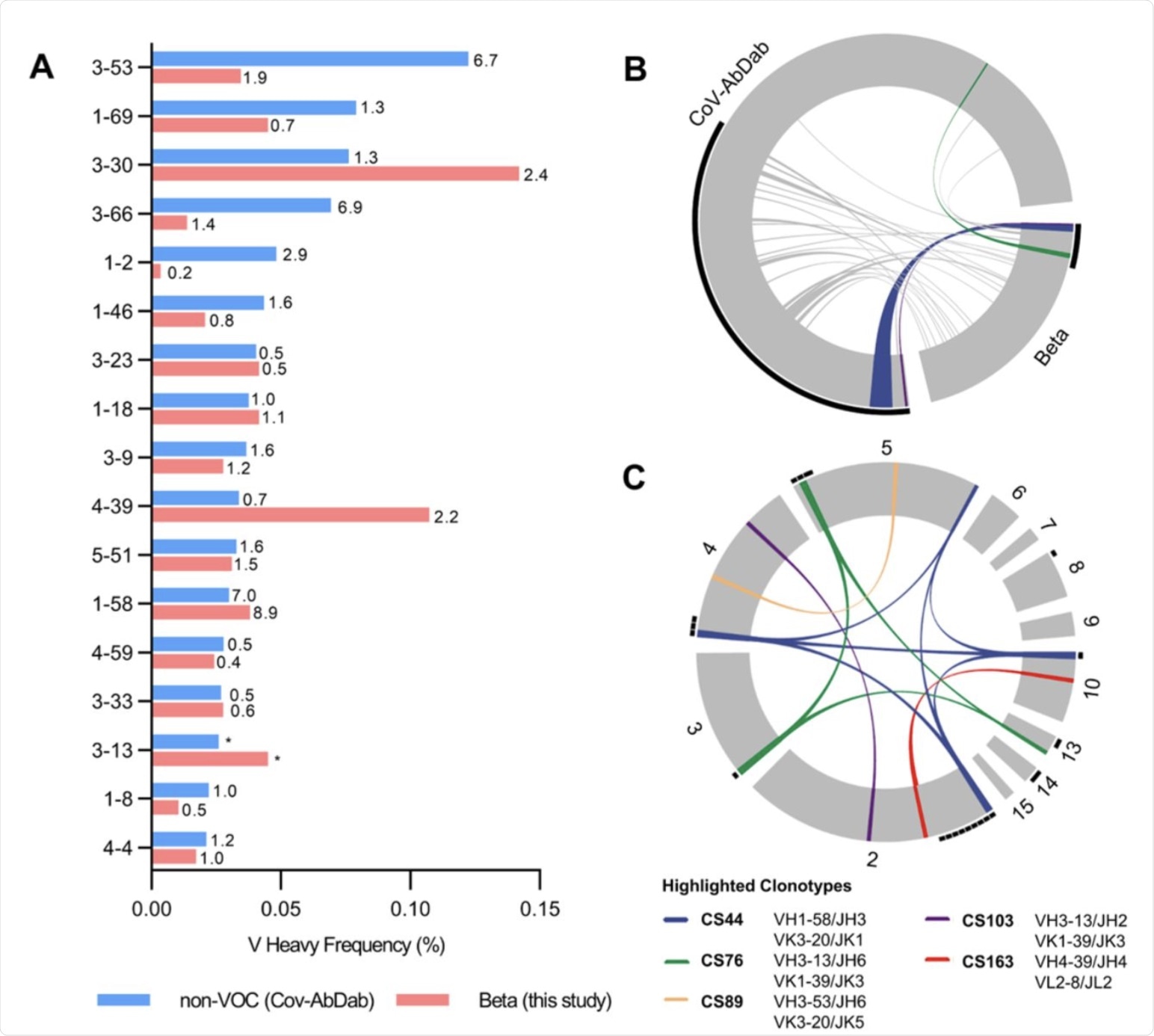 Germline gene usage and clonotype analysis of Beta-elicited antibodies. (A) VH gene usage of 289 RBD Beta IgG mAbs from this study (red) is compared to 1037 non-VOC RBD mAbs from 96 previously published studies (blue, Cov-AbDab) (17). Frequencies of mAbs encoded by each VH gene are shown as bars. Enrichment of indicated VH genes is compared to healthy individuals (31) with fold-enrichment shown as number next to bars. VH gene frequencies that were not reported in healthy individuals (31) are shown with asterisk (*). Only VH genes with a frequency of at least 2% in Cov-AbDab are shown and VH genes are ordered by frequency in Cov-AbDab. (B) Circos plot shows the relationship between 289 IgG mAbs from this study (Beta) and 1037 previously published human mAbs reactive to RBD (non-VOC) from 96 studies (17). Interconnecting lines display clonotypes shared between both datasets, as defined by the usage of the same V and J gene on both Ig heavy and light chain. Thin black lines at the outer circle border indicate expanded clonotypes within the respective data set. (C) Circos plot displaying the 289 IgG mAbs from this study grouped per patient. Interconnecting colored lines indicate clonotypes found in more than one patient. Small black angles at the outer circle border indicate clonally expanded clones within one patient. (B-C) Colored interconnecting lines depict clonotypes found in more than one patient of our cohort.
Germline gene usage and clonotype analysis of Beta-elicited antibodies. (A) VH gene usage of 289 RBD Beta IgG mAbs from this study (red) is compared to 1037 non-VOC RBD mAbs from 96 previously published studies (blue, Cov-AbDab) (17). Frequencies of mAbs encoded by each VH gene are shown as bars. Enrichment of indicated VH genes is compared to healthy individuals (31) with fold-enrichment shown as number next to bars. VH gene frequencies that were not reported in healthy individuals (31) are shown with asterisk (*). Only VH genes with a frequency of at least 2% in Cov-AbDab are shown and VH genes are ordered by frequency in Cov-AbDab. (B) Circos plot shows the relationship between 289 IgG mAbs from this study (Beta) and 1037 previously published human mAbs reactive to RBD (non-VOC) from 96 studies (17). Interconnecting lines display clonotypes shared between both datasets, as defined by the usage of the same V and J gene on both Ig heavy and light chain. Thin black lines at the outer circle border indicate expanded clonotypes within the respective data set. (C) Circos plot displaying the 289 IgG mAbs from this study grouped per patient. Interconnecting colored lines indicate clonotypes found in more than one patient. Small black angles at the outer circle border indicate clonally expanded clones within one patient. (B-C) Colored interconnecting lines depict clonotypes found in more than one patient of our cohort.
The authors also determined the residues that establish the mAb binding selectivity for the 37 RBD Beta-specific mAbs via enzyme-linked immunosorbent assay (ELISA). For the three RBD mutations that define the Beta variant (K417N, E484K, and N501Y), the authors identified mAbs with RBD binding that was dependent on a single residue, while the RBD specificity of other mAbs was dependent on multiple residues.
A broad variety of VH genes encode for RBD Beta-specific mAbs. All VH4-39 mAbs with RBD Beta specificity from three different participants were all dependent on Y501, which is 81.8% of the mAbs within the specific category.
From this finding, it could be suggested that there is a common binding mechanism of these mAbs that is dependent on Y501, a mutation found in RBD Alpha, Beta, and Gamma, but not Delta. This observation may explain the frequent use of VH4-39 in mAbs to RBD Beta.
Few somatic hypermutations were revealed by VH4-39 Y501 dependent mAbs in VH genes; however, no uniform pattern in other sequence features was observed. Despite all VH4-39 RBD Beta-specific mAbs binding to a Y501 dependent epitope, there are notable differences in their neutralization activity against authentic Beta virus.
Implications
The findings from this study provide important insights that could be considered for next-generation vaccine design and antibody-based therapeutics against current SARS-CoV-2 VOCs and any future SARS-CoV-2 variants that may arise. For example, vaccination programs based on various RBD sequences could be analyzed for superiority in the induction of cross-variant immunity, including recruitment of highly potent VH1-58 antibodies.
Promising cross-variant antibody titers have already been shown in novel vaccine candidates in pre-clinical trials that are based on the Beta variant. It could be suggested that, alongside these clinical trials, studies with the aim of characterizing the immune response regarding further variants should be conducted, especially including the Delta variant, which is currently the dominating VOC globally.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Reincke, S. M., Yuan, M., Kornau, H., et al. (2021). SARS-CoV-2 Beta variant infection elicits potent linage-specific and cross-reactive antibodies. biorxiv. doi:10.1101/2021.09.30.462420. https://www.biorxiv.org/content/10.1101/2021.09.30.462420v1.
Posted in: Molecular & Structural Biology | Microbiology | Medical Science News | Medical Research News | Medical Condition News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Assay, Cell, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Frequency, Gene, Genes, Germline, Immune Response, immunity, Mutation, Pandemic, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Therapeutics, Vaccine, Virus
.jpg)
Written by
Colin Lightfoot
Colin graduated from the University of Chester with a B.Sc. in Biomedical Science in 2020. Since completing his undergraduate degree, he worked for NHS England as an Associate Practitioner, responsible for testing inpatients for COVID-19 on admission.
Source: Read Full Article
